Research
Research Overview
- Research Program Overview
EMRI RESEARCH PROGRAM
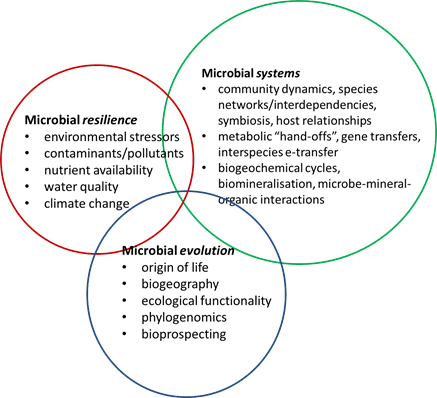 EMRI research is grouped into three interconnected themes of microbial resilience, microbial systems and microbial evolution. The three themes underpin the research program that will address five key research objectives:
EMRI research is grouped into three interconnected themes of microbial resilience, microbial systems and microbial evolution. The three themes underpin the research program that will address five key research objectives:- Contribute strategically to the growing global catalogue of environmental genomes and metagenomics datasets
- Understand key environmental factors that shape microbial communities
- Predict adaptations in microbial communities and their functionality under changing environmental conditions
- Identify novel metabolic/enzymatic pathways for nutrient cycling or contaminant remediation
- Use metagenomics to drive cultivation-based hypothesis-testing experiments and selective enrichment/species isolations.
Research Projects
Geomicrobiology
- Geomicrobiology and biogeochemistry of mercury methylation
Geomicrobiology and biogeochemistry of mercury methylation
 The Moreau Geomicrobiology Lab studies the processes by which certain microorganisms convert toxic ionic mercury (Hg2+) into the more toxic organometallic compound, methylmercury (CH3Hg+).
The Moreau Geomicrobiology Lab studies the processes by which certain microorganisms convert toxic ionic mercury (Hg2+) into the more toxic organometallic compound, methylmercury (CH3Hg+).This process occurs in aqueous environments from riverine sediments to the open ocean, but the mechanisms and triggers for microbial mercury methylation are still poorly understood.
- Bioremediation of mining impacted fresh water
Bioremediation of mine wastes
Gold mining produces toxic heavy metals and other chemical waste products.

Some environmental microorganisms can biodegrade these contaminants into less harmful or non-toxic substances.
The Moreau Geomicrobiology Lab applies both cultivation-based and molecular biology approaches to developing new biotechnologies for mitigating mining impacts on soil and groundwater
Plant and Soil Microbiology
- Understanding the transmission of antibiotic resistance in agro-ecosystems
Funded by the ARC – DP170103628, $399,800
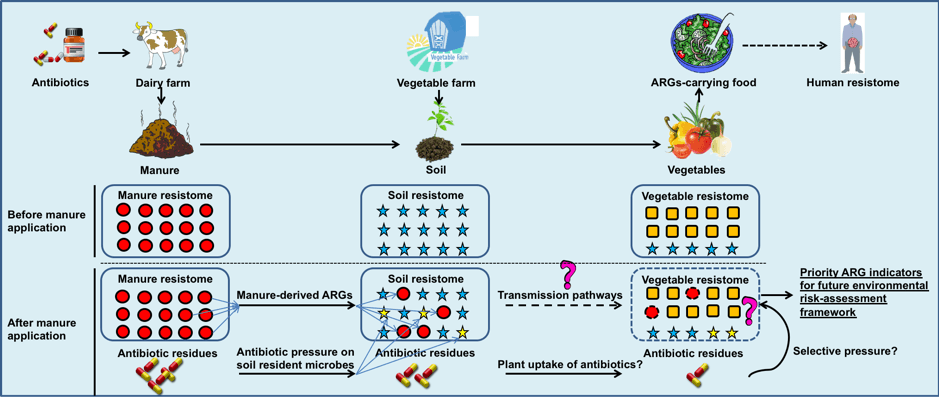 The emerging spread of antibiotic resistance genes (ARGs) represents a serious threat to human health. However, we have very limited knowledge about the pathways and mechanisms for transmission of ARGs in the environment. This project will use advanced molecular approaches to investigate all the major classes of ARGs in typical vegetable farms and animal manure, and to explore the major routes for transmission of ARGs from manure to soil and from rhizosphere to the vegetable surfaces and endophytic bacterial communities. The results will be integrated to identify the priority ARGs indicators with high likelihood of spread into food chain, and contribute to the development of management options to tackle environmental antibiotic resistance.
The emerging spread of antibiotic resistance genes (ARGs) represents a serious threat to human health. However, we have very limited knowledge about the pathways and mechanisms for transmission of ARGs in the environment. This project will use advanced molecular approaches to investigate all the major classes of ARGs in typical vegetable farms and animal manure, and to explore the major routes for transmission of ARGs from manure to soil and from rhizosphere to the vegetable surfaces and endophytic bacterial communities. The results will be integrated to identify the priority ARGs indicators with high likelihood of spread into food chain, and contribute to the development of management options to tackle environmental antibiotic resistance.Project team: Jim He (Project leader), Hangwei Hu (Lead researcher), and external partners
- Soil microbial indicators for efficient use of nitrification inhibitors
Funded by the ARC – LP160101134, $237,000
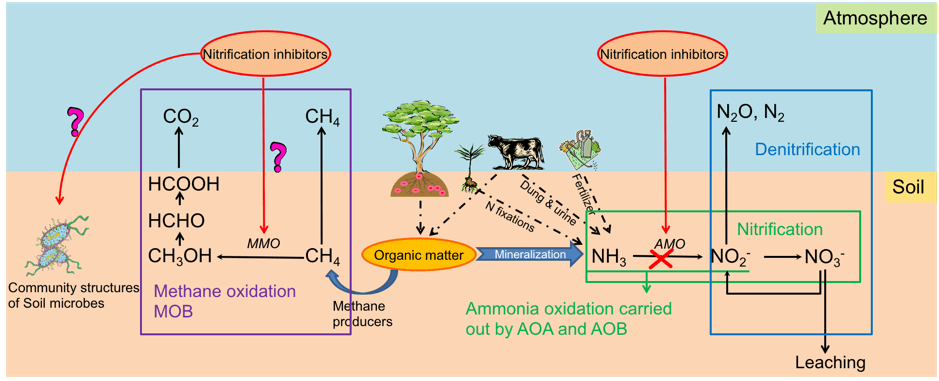 Nitrification inhibitors are one tool widely used to improve nitrogen fertiliser efficiency and reduce greenhouse gas nitrous oxide emissions. However their effectiveness is variable across soil types and one possible reason is the different microbial communities that exist in these soils. The project addresses the key knowledge gaps of interactions between the nitrification inhibitors and the soil functional microbial communities. The project aims to fundamentally improve our understanding of the efficiency and governing factors of the nitrification inhibitors in different agricultural soils, and thus provide guidance to develop sound management strategies to improve fertiliser nitrogen use efficiency in Australian Agricultural soils.
Nitrification inhibitors are one tool widely used to improve nitrogen fertiliser efficiency and reduce greenhouse gas nitrous oxide emissions. However their effectiveness is variable across soil types and one possible reason is the different microbial communities that exist in these soils. The project addresses the key knowledge gaps of interactions between the nitrification inhibitors and the soil functional microbial communities. The project aims to fundamentally improve our understanding of the efficiency and governing factors of the nitrification inhibitors in different agricultural soils, and thus provide guidance to develop sound management strategies to improve fertiliser nitrogen use efficiency in Australian Agricultural soils.Project team: Jim He (Project leader), Helen Suter, Deli Chen and external partners.
- Reserving nitrogen in soils through microbial nitrate reduction to ammonium
Funded by the ARC – DP160101028, $248,000
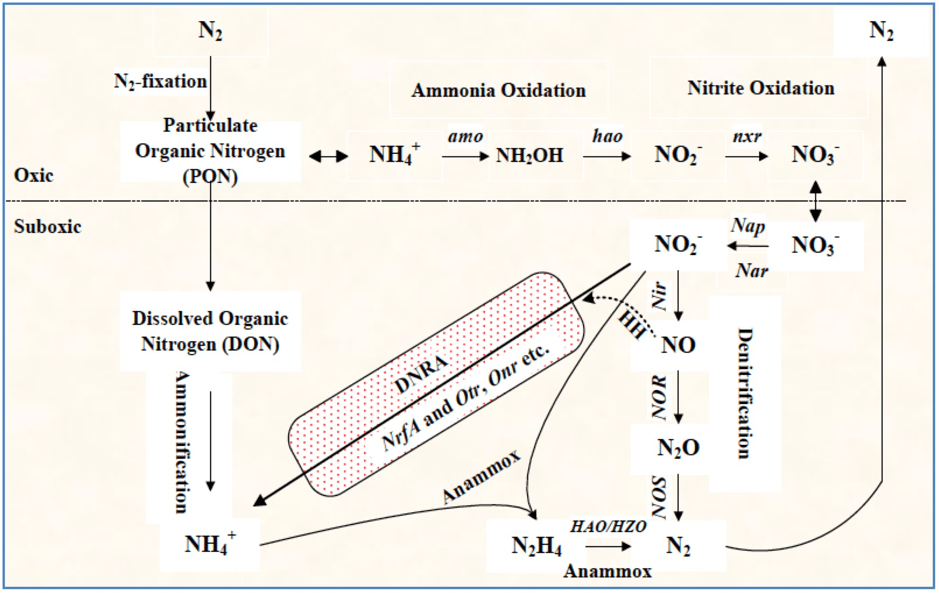 This project aims to identify those microbes able to transform nitrate to ammonium and thus increase soil nitrogen conservation. More than 50 per cent of the nitrogen in fertilisers applied to soils is lost into the environment, which is both a financial loss to farmers and a main anthropogenic source of nitrogen pollution. Some microbes can transform nitrate into ammonium through dissimilatory reduction (DNRA) and thus increase soil nitrogen retention. However, the DNRA process and the responsible microbial groups remain largely unknown. This project plans to use isotope tracing and biomolecular approaches to identify those DNRA microbial groups and elucidate the DNRA reaction process. The findings may support the use of DNRA to improve soil nitrogen.
This project aims to identify those microbes able to transform nitrate to ammonium and thus increase soil nitrogen conservation. More than 50 per cent of the nitrogen in fertilisers applied to soils is lost into the environment, which is both a financial loss to farmers and a main anthropogenic source of nitrogen pollution. Some microbes can transform nitrate into ammonium through dissimilatory reduction (DNRA) and thus increase soil nitrogen retention. However, the DNRA process and the responsible microbial groups remain largely unknown. This project plans to use isotope tracing and biomolecular approaches to identify those DNRA microbial groups and elucidate the DNRA reaction process. The findings may support the use of DNRA to improve soil nitrogen.
- Fungal Biodiversity
 The fungal kingdom are likely the most species rich group of microbes on earth, yet we have described less than 3% of all estimated species, and have only a limited understanding of how fungi contribute to the many environments in which they are found. From the species that have been studied, it is clear that fungi are critical for nutrient cycling and in interaction with plants and other microbes.
The fungal kingdom are likely the most species rich group of microbes on earth, yet we have described less than 3% of all estimated species, and have only a limited understanding of how fungi contribute to the many environments in which they are found. From the species that have been studied, it is clear that fungi are critical for nutrient cycling and in interaction with plants and other microbes.We are interested in fungal diversity from two perspectives. First is the impact of fungi and other microbes on disease in agricultural settings, in our case how other microbes impact the major diseases that impact the Australian canola (Brassica napus) industry. Second, is with colleagues at the Royal Botanic Gardens Victoria we are investigating the diversity and distribution of some of the poorly understood fungal taxa.
Contact: Dr Alexander Idnurm at the Mycology Lab
Marine Microbiology
- Manipulation of microbes to assist coral adaptation to climate change
Funded by the ARC – DP160101468, $494,700 (van Oppen and Blackall) and FL180100036, $3,011,916 (van Oppen)
 Our group is called the Marine Microbial Symbiosis group. We explore how corals interact with their associated microbial communities, and attempt to augment the capacity of corals to tolerate stress by manipulating these communities. Our research aims to support development of novel coral reef restoration approaches.
Our group is called the Marine Microbial Symbiosis group. We explore how corals interact with their associated microbial communities, and attempt to augment the capacity of corals to tolerate stress by manipulating these communities. Our research aims to support development of novel coral reef restoration approaches.We use the sea anemone Exaiptasia pallida as a model animal, and focus on the role of prokaryotes and the algal symbiont Symbiodinium. The approaches used to manipulate microbial communities include use of probiotics, experimental evolution and genetic engineering.
See our Recent publication: Damjanovic K, Blackall LL, Webster NS, van Oppen MJH (2017) The contribution of microbial biotechnology to mitigating coral reef degradation. Microbial Biotechnology, doi:10.1111/1751-7915.12769.
Contacts: Laureate Fellow Prof Madeleine van Oppen @ Google Scholar and EMRI Director Prof Linda Blackall @ Google Scholar.
Photo: Australian Institute of Marine Science.
- Fluid dynamics of corals
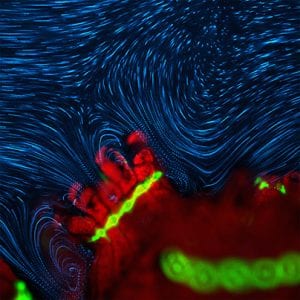 Fundamental processes underpinning the health and function of corals such as nutrient uptake, oxygen exchange and bacterial infection occur at the microscale, yet we still largely lack a quantitative understanding of the governing transport processes. The cilia that corals use for feeding and cleansing also generate dramatic vortical flows, which affect mass transport and may provide a barrier against infection. My research investigates the nature and consequences of these active vortical flows. I use state of the art visualisation of flows, tracking of individual bacterial cells, and mathematical modelling to quantitatively understand the coupling between ciliary flows, nutrient and oxygen dynamics, and pathogen behaviour.
Fundamental processes underpinning the health and function of corals such as nutrient uptake, oxygen exchange and bacterial infection occur at the microscale, yet we still largely lack a quantitative understanding of the governing transport processes. The cilia that corals use for feeding and cleansing also generate dramatic vortical flows, which affect mass transport and may provide a barrier against infection. My research investigates the nature and consequences of these active vortical flows. I use state of the art visualisation of flows, tracking of individual bacterial cells, and mathematical modelling to quantitatively understand the coupling between ciliary flows, nutrient and oxygen dynamics, and pathogen behaviour.
- Micro-niches in the coral skeleton
Coral reefs are spectacular ecosystems held together by calcium carbonate structures secreted by corals (their skeletons). The diversity, spatial structure and functions of the microbiota residing in the skeleton underneath the coral tissue are barely understood.

We study the coral skeleton microbiome, focusing on the interactions between microbiota and the steep environmental gradients in oxygen, light and other nutrients present in the skeleton. We use planar optode imaging to visualise gradients in physicochemical properties and characterise of the microbial communities at different positions along the environmental gradients using molecular techniques.
Contact: Heroen Verbruggen at Google Scholar
- Endosymbiosis in the algal world
Microbial endosymbiosis, i.e. symbiosis where one cell lives inside another cell, is very commonly found in nature. These partnerships range from transient, fairly loose associations to well-established symbioses where neither of the partners can live without the other.
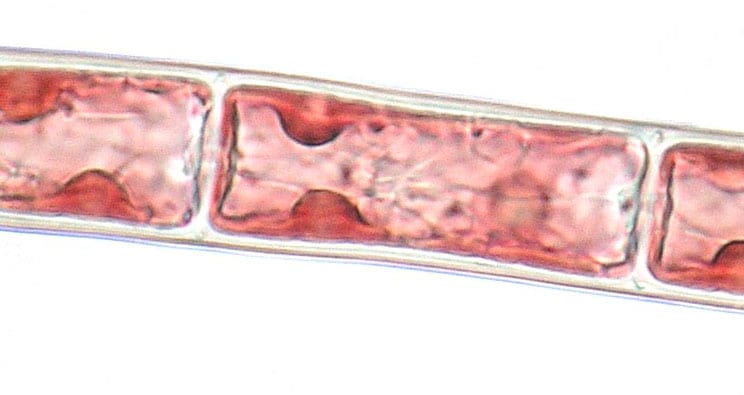
We are interested in many aspects of microbial symbiosis in marine algae. We characterise the nature and evolution of bacterial endosymbionts living within seaweeds and the origins of algal chloroplasts through endosymbiosis. We use a combination of microscopy, metabarcoding and whole-genome sequencing to characterise the partners, how they interact, and how their associations have evolved through geological time.
Contact: Heroen Verbruggen at Google Scholar
- Microbiologically influenced corrosion
Microbiologically influenced corrosion
 Accelerated low water corrosion (ALWC) is a type of microbiologically influenced corrosion that substantially increases damage to metal structures in seawater around the low tide level. ALWC is a costly global issue.
Accelerated low water corrosion (ALWC) is a type of microbiologically influenced corrosion that substantially increases damage to metal structures in seawater around the low tide level. ALWC is a costly global issue.We hypothesise that the functional features of microbes, including their propensity to attach to metal surfaces and their metabolic pathways, facilitate ALWC. We aim to identify the microbes in ALWC tubercles with the aim of better understanding this phenomenon.
A recent paper: In Microbiology Australia, September 2018. Development of a laboratory test for microbial involvement in accelerated low water corrosion. By Scott Wade and Linda Blackall
Contact: Linda Blackall @ Google Scholar
Food Microbiology
- Microbial diversity and interactions in food ecosystems: from model to meal
 Why does food taste better when a community of microbes is present? What can interactions between microorganisms tell us about wider ecological systems? The Howell lab studies yeast and bacterial communities in a wide variety of food ecosystems, including bread, beer, wine and chocolate. Analytical methods help us to understand the biochemical pathways of yeasts and bacteria that influence flavour production and draws these together with broader ecological and social understandings of taste, health and sustainability.
Why does food taste better when a community of microbes is present? What can interactions between microorganisms tell us about wider ecological systems? The Howell lab studies yeast and bacterial communities in a wide variety of food ecosystems, including bread, beer, wine and chocolate. Analytical methods help us to understand the biochemical pathways of yeasts and bacteria that influence flavour production and draws these together with broader ecological and social understandings of taste, health and sustainability.Contact: Dr Kate Howell
Computational Statistics and Microbiology
- Development and application of multivariate methods for microbiome studies
There are major statistical and computational challenges in analysing microbial communities that currently hinder the potential of microbiome research to substantially advance biomedical understanding. We are currently expanding mixMC to better characterise and understand important microbiome-host interactions. Some of our methods developments aim at addressing batch effects in microbiome experiments and analyse scarce temporal sampling in time course studies.
We analyse microbiome datasets from our collaborators for a wide range of studies, including investigating the role of gut and oral microbiome in spondyloarthropathy diseases, the development of intestinal or salivary microbiota in toddlers and infants, investigating the gut-brain crosstalk in Huntington’s disease.
 Contact: NHMRC Career Development Fellow Dr Kim-Anh Lê Cao @ Google Scholar
Contact: NHMRC Career Development Fellow Dr Kim-Anh Lê Cao @ Google ScholarSee a publication by Kim-Anh:
MixMC: A Multivariate Statistical Framework to Gain Insight into Microbial Communities
- Development of the mixOmics R toolkit package
mixOmics is one of the few
 R package dedicated to the integration of multiple ‘omics data (19 novel methodologies implemented so far, amongst which 13 were developed by our lab) and with an increasing uptake from the research community. The package has been downloaded > 29K times in 2017, (R CRAN package download logs). Programming developments are on-going for interactive web interfaces, and efficient programming for large-scale studies. Check our our recent publication (Rohart et al. 2017b) and a poster that gives an overview of this large project. The mixOmics team run multiple day workshops for an introduction to multivariate projection-based methods for data integration using mixOmics, see our website www.mixOmics.org for news and tutorials.
R package dedicated to the integration of multiple ‘omics data (19 novel methodologies implemented so far, amongst which 13 were developed by our lab) and with an increasing uptake from the research community. The package has been downloaded > 29K times in 2017, (R CRAN package download logs). Programming developments are on-going for interactive web interfaces, and efficient programming for large-scale studies. Check our our recent publication (Rohart et al. 2017b) and a poster that gives an overview of this large project. The mixOmics team run multiple day workshops for an introduction to multivariate projection-based methods for data integration using mixOmics, see our website www.mixOmics.org for news and tutorials.Contact: NHMRC Career Development Fellow Dr Kim-Anh Lê Cao @ Google Scholar – See a publication by Kim-Anh:
mixOmics: An R package for ‘omics feature selection and multiple data integration
Mathematical Microbiology
- Microbial fluid dynamics
Microbial fluid dynamics
 Fluid mechanics at the microscale governs a myriad of physical, chemical and biological processes in the environment.
Fluid mechanics at the microscale governs a myriad of physical, chemical and biological processes in the environment.We apply advanced video-microscopy and microfluidic techniques to directly visualise dynamic processes at the single cell level, from the collective beating of cilia to the active search behaviour of motile bacteria.
These results will uniquely inform the development of mathematical models with far-reaching ecological insights.
Contact: DECRA Fellow Dr Douglas Brumley @ Google Scholar

